HOLLOW
phosphate channel in OprP
The OprP porin [2O4V] allows the transport of phosphate across the outer membrane of a bacteria. It is expressed under low phosphate conditions that allows for the passage of phosphate. To illustrate the channel of this porin, we only look at chain A, so in the pdb file, we delete all the other chains and save this in '2o4v-a.pdb'.

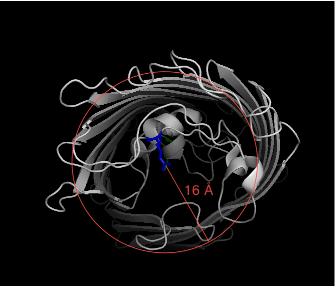
In order to construct a surface of the open-ended channel, we need to define a cylinder around the protein where the intersection of the cylinder with the protein defines the channel opening. We need to choose a radius for the cylinder. It doesn't matter if the radius extends out of the body of the protein:
This information will be stored in the 'constraints' file as the lines:
'remove_asa_shell': False,
'type': 'cylinder',
'radius': 16.0,
To define the cylinder, we need to choose 2 atoms to define the central axis of the cylinder. It doesn't matter if the atoms are above or below the length of the cylinder as we can use offsets to define the extent of the cylinder.
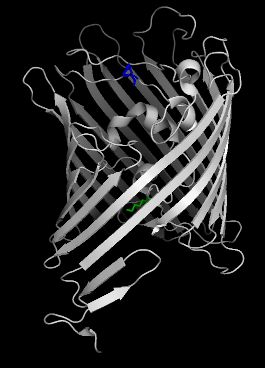
For the bottom atom A-LYS-318-CD (green), we put in the 'constraint' file where the offset of 5.0 � means the cylinder will extend out out below the green residue by 5.0 �:
'chain1': 'A',
'res_num1': 318,
'atom1': 'NZ',
'axis_offset1': 5.0,
For the top atom A-LYS-318-NZ (blue), the offset of -5.0 � means the cylinder will contract 5.0 � below the blue atom:
'chain2': 'A',
'res_num2': 218,
'atom2': 'CD',
'axis_offset2': -5.0,
Putting this all together, we type in a text file called 'constraint':
{
'remove_asa_shell': False,
'type': 'cylinder',
'radius': 16.0,
'chain1': 'A',
'res_num1': 318,
'atom1': 'NZ',
'axis_offset1': 5.0,
'chain2': 'A',
'res_num2': 218,
'atom2': 'CD',
'axis_offset2': -5.0,
}
We now run python hollow with the constraint file that will generate the hollow spheres based on this constraint, with the important information that hollow spheres on the surface of the cylinder are labeled occupancy q=0, and hollow spheres inside the cylinder are labeled occupancy q=1:
>>> hollow -c constraint -o hollow.pdb 2o4v-a.pdb
We now load the hollow spheres load into pymol into the objects "hollow" and "2o4v-a". Notice the cylinder extends past the green residue (axis_offset1 = 5 �) but doesn't reach the blue residue (axis_offset2 = -5 �)
>>> pymol hollow.pdb 2o4v-a.pdb
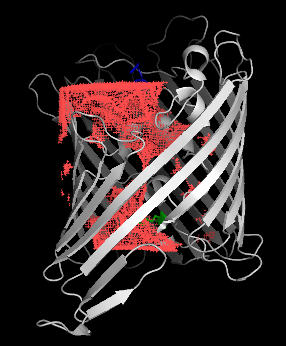
Once in pymol, we show the channel surface by showing the surface of hollow spheres inside the cylinder (q=1) using two-sided lighting:
pymol> show surface, hollow and q>0
pymol> hide nonbonded
pymol> set two_sided_lighting, on
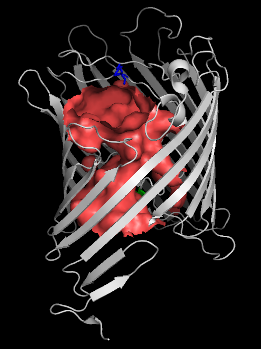
Using the hollow spheres, choosing the channel residues is trivial:
pymol> select lining, byres hollow around 5
pymol> show sticks, lining
pymol> cartoon tube
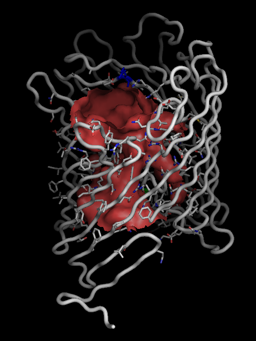
And we can get a very detailed picture of the channel-lining residues with the channel:
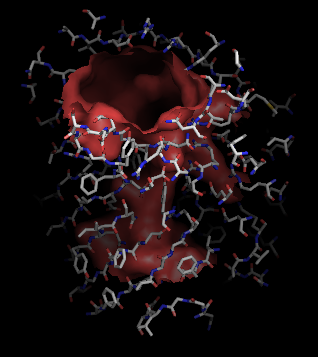
This process can be repeated using different grid-spacings for more accurate surfaces. The size and orientation of the cylinder can be adjusted by varying the radius, and choice of anchor atoms and offsets. Changing the cylinder will allow different choices in showing the channel opening.
Coloring by electrostatic potential
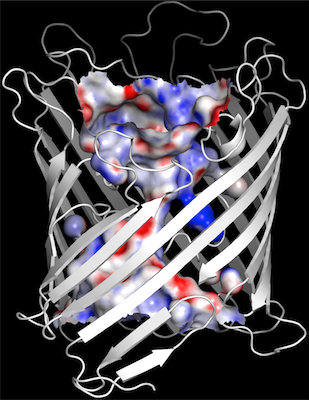
-
to generate the electrostatic potential, we prefer the online tool
pdb2pqr
because PYMOL's hydrogen assignments are not that great. We use the
PARSE forcefield w/ default settings + PROPKA assigned at pH7,
add/keep chain IDs, and neutral termini if the termini are
disordered and missing from the model. Examine the propka file and
save this and the pqr file.
-
From the same webpage launch APBS online. Use custom values if you
wish. Save and unzip the dx file.
-
Load the dx file and hollow pdb file into pymol. Launch the APBS
tool and go to the visualization tab. Select the map and the hollow
object as the molecule and update. Set the molecular surface
potentials (We use -15 to -20 low, 15 to 20 high) then click update.
-
Open the channel surface by selecting q=0 for the hollow object and
then hide surface.