HOLLOW
oxygen pathways in myoglobin
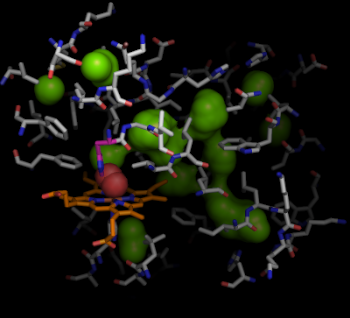
Myoglobin [1MBO] (grey) binds oxygen (red) through a heme group (beige). The binding site of oxygen is accessible through the gate at His64 (purple), which swings open. The pathways that oxygen can take through the interior of myoglobin are important for understanding the function of myoglobin.
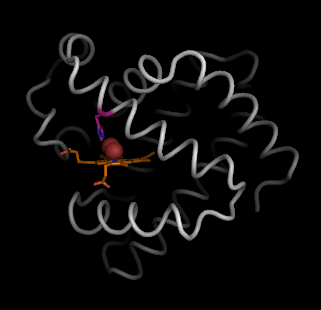
To illustrate the pathways of myoglobin, we generate the voids by applying HOLLOW to myoglobin using the default grid-spacing 0.25 � in the automated mode.
>>> hollow -o hollow.pdb -g 0.25 1mbo.pdb
To view the structure with the hollow spheres, fire up Pymol. Pymol takes a bit of work to learn but is very powerful.
>>> pymol 1mbo.pdb hollow.pdb
This will load two objects into Pymol: "1mbo" and "hollow". To start with, show the hollow spheres in the typical "nonbonded" cross-hair display (green).
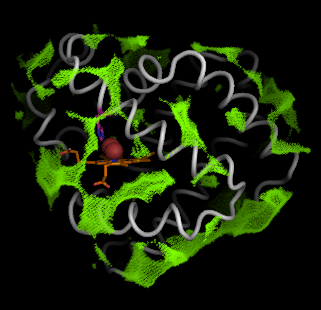
First, let's look at the hollow object. Select individual hollow spheres clustered in the interior, and rename this selection as the "seed". You only need to select one hollow sphere in a cluster.
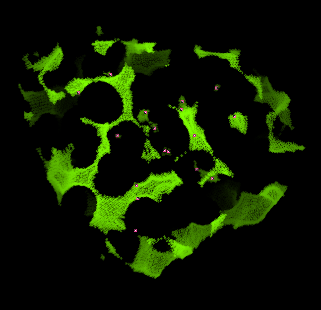
Using the pymol command expand, we can now select entire clusters found in the interior of the protein:
pymol> select inner, hollow within 4 of seed
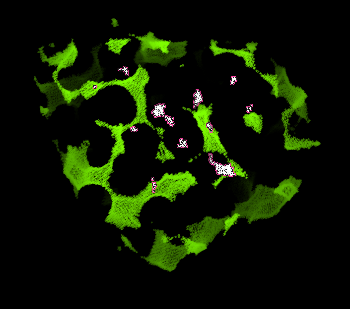
We remove atoms that are not in the interior, i.e. atoms not part of the "inner" selection.
pymol> remove hollow and not inner
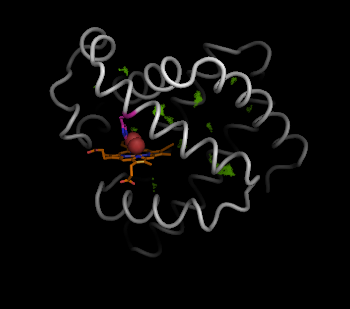
Now we have the interior hollow spheres, we can show this clearly using the sphere view:
pymol> show sphere, inner
pymol> set sphere_quality, 2
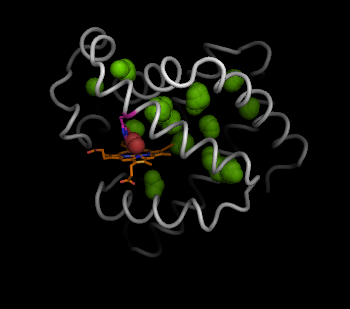
However, a better way to view the negative space described by the hollow spheres is to draw the connected surface of these interior hollow spheres.
pymol> show surface, inner
pymol> set surface_quality, 2
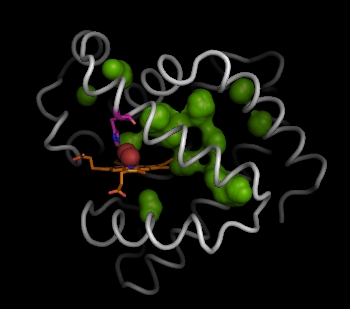
Finally, we can use the hollow spheres to easily, and simply select the residues that line the voids and pathways inside myoglobin. This can be simply done by selecting residues of myoglobin (1mbo) in proximity to the hollow spheres:
pymol> select lining, byres inner around 5
pymol> show sticks, lining
pymol> hide everything, 1mbo and not lining